Lab 5, Ioan Dragomir
Process Modeling, prof. Daniel Moga, Technical University of Cluj-Napoca
Piecewise linear approximations of measured functions.
Contents
Intro
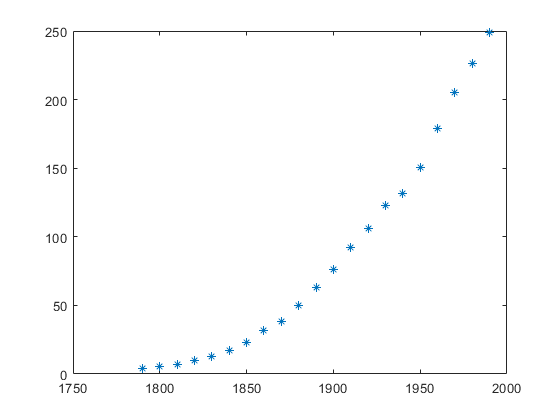
Census example
load census; x = cdate; y = pop; plot(x, y, '*');
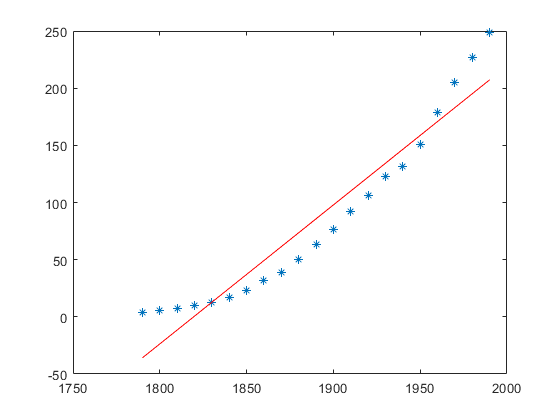
f = fit(x, y, 'poly1') p = coeffvalues(f) a = p(1) b = p(2) y_approx = a * x + b; hold on; plot(x, y_approx, 'r');
f =
Linear model Poly1:
f(x) = p1*x + p2
Coefficients (with 95% confidence bounds):
p1 = 1.216 (1.045, 1.387)
p2 = -2212 (-2535, -1889)
p =
1.0e+03 *
0.0012 -2.2120
a =
1.2157
b =
-2.2120e+03
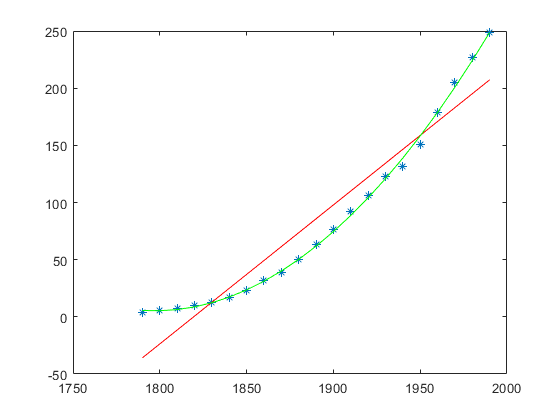
f = fit(x, y, 'poly2'); p = coeffvalues(f); a = p(1); b = p(2); c = p(3); y_approx_2 = a * x.^2 + b * x + c; plot(x, y_approx_2, 'g');
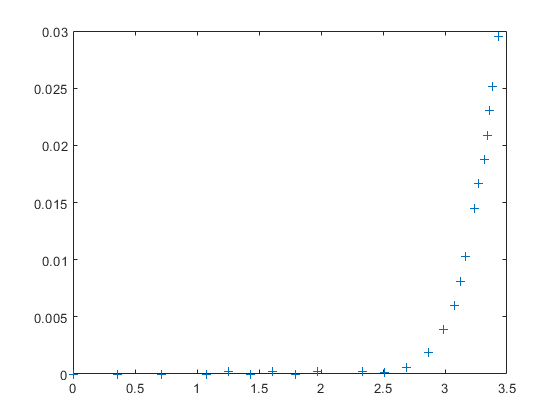
LED
IV characteristic for an ideal LED is a sudden jump to \infty after the threshold voltage.
Measuring a real LED, we get values like these:
I_measured = [0, 0, 0, 0, 0.0002, 0, 0.0002, 0, 0.0002, 0.0002, 0.0001, ... 0.0006, 0.00189, 0.00389, 0.00598, 0.00807, 0.01027, 0.01445, ... 0.01664, 0.01874, 0.02083, 0.02302, 0.02512, 0.02950]'; V_measured = [0, 0.36091, 0.71343, 1.07434, 1.25060, 1.43525, 1.61151, ... 1.79616, 1.97242, 2.33333, 2.50959, 2.68585, 2.87050, 2.98801, ... 3.08034, 3.12230, 3.16427, 3.23981, 3.27338, 3.31535, 3.34053, ... 3.35731, 3.38249, 3.43285]'; clf; plot(V_measured, I_measured, '+');
Which we may try to fit with a single linear function:
f = fit(V_measured, I_measured, 'poly1'); p = coeffvalues(f); a1 = p(1); b1 = p(2); I_predicted1 = a1 * V_measured + b1; clf; hold on; plot(V_measured, I_measured, '+'); plot(V_measured, I_predicted1, 'r');
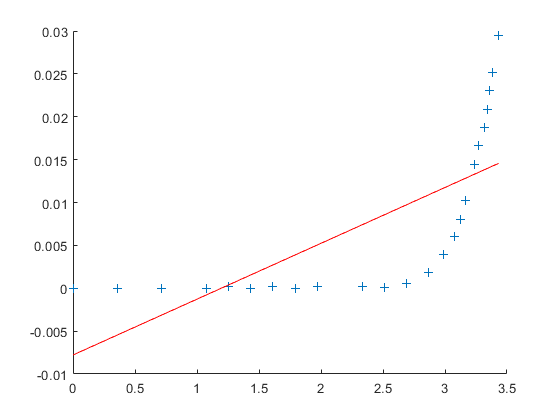
But this is very clearly a rather disappointing model.
Piecewise linear model
We may then attempt to model the IV curve by a set of linear pieces. The exercise mentions using 5 intervals, but not which data points each one covers, so we must decide that ourselves. After some attempts, the cleanest division I came across is:
num_intervals = 5; points_per_interval = [11, 3, 3, 6, 5];
Which can then be converted to a list of "interval boundaries" with the following code. Reverse engineering not really relevant, it just adds them in a slightly special way to allow for a one point overlap
assert(length(points_per_interval) == num_intervals);
assert(sum(points_per_interval) - num_intervals + 1 == length(V_measured));
indices = [0 cumsum(points_per_interval)] + 1 - (0:5); %Voodoo magic
I_predicted2 = zeros([1, length(V_measured)]); for i = 1 : 5 i0 = indices(i); i1 = indices(i+1); V0 = V_measured(i0); V1 = V_measured(i1); f = fit(V_measured(i0:i1), I_measured(i0:i1), 'poly1'); p = coeffvalues(f); a = p(1); b = p(2); I_predicted2(2*i-1) = a * V0 + b; I_predicted2(2*i) = a * V1 + b; plot([V0, V1], I_predicted2(2*i-1:2*i), 'g-'); end x0, x1 = xlim; y0, y1 = ylim; for x = indices plot([x, x], [y0, y1], 'k--'); end xlim([x0, x1]); ylim([y0, y1]);
Unrecognized function or variable 'x0'. Error in lab5 (line 128) x0, x1 = xlim;